New paper - A quantitative model of enzyme-free copying of RNA with dimers
Burger et al., Nucleic Acids Research (2025)
20.10.2025
Ludwig Burger, Franziska Welsch, Eric Kervio, Marc Henker, Gabrielle Leveau, Ulrich Gerland and Clemens Richert
Nucleic Acids Research doi.org/10.1093/nar/gkaf987
Abstract
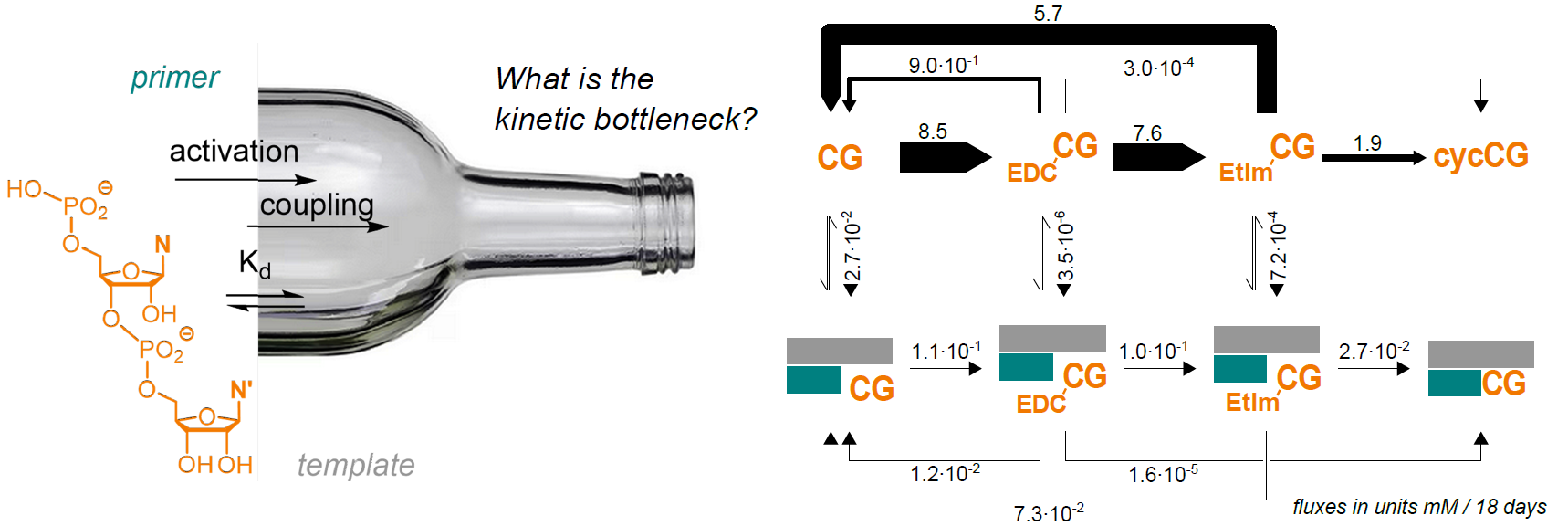
The transfer of genetic information from one RNA strand to a daughter strand in the absence of polymerases could have initiated prebiotic evolution. This reaction, driven by base pairing and chemical reactivity alone, is slow and low yielding. The molecular basis of its inefficiency has remained unclear. We conducted a systems chemistry analysis of the most effective enzyme-free RNA copying system to date. This system uses the strongly pairing dimers (CC, CG, GC, and GG) and in situ activation. Rate constants for activation were obtained from nuclear magnetic resonance-monitored model reactions, effective dissociation constants in the sub-millimolar range were obtained from inhibitor assays, and rates of phosphodiester formation for bound and activated dimers were determined by fitting extension kinetics, with the latter ranging from 1.4–16 × 10−3 h−1. Using a kinetic model that incorporates all experimentally determined parameters, we simulated primer extension. This identified phosphodiester formation on templates as the rate-limiting step. Our model shows copying of up to 12 template bases through a combination of primer extension, dimer–dimer coupling, and fragment ligation steps. The fluxes through reaction channels provide an unprecedented view of genetic copying in a protein-free RNA system. Our analysis identifies the remaining bottlenecks of enzyme-free pathways and provides the basis for rationally searching for more efficient self-replicating systems.